---
title: Comparing Experiments
---
It is always useful to be able to do some forensics on what causes an experiment to succeed and to better understand
performance issues.
The **ClearML Web UI** provides a deep experiment comparison, allowing to locate, visualize, and analyze differences including:
* [Details](#details)
- [Artifacts](#artifacts) - Input model, output model, and model design.
- [Execution](#execution-details) - Installed packages and source code.
- [Configuration](#configuration) - Configuration objects used by the experiment.
* [Hyper Parameters](#hyperparameters)
- [Values (table) view](#values-mode) - Key/value of all the arguments used by the experiments.
- [Parallel coordinates view](#parallel-coordinates-mode) - Impact of each argument on a selected metric
the experiments reported (see [task.connect_configuration](../references/sdk/task.md#connect_configuration)).
* [Scalars](#scalars)
- Specific values and plots of scalar series (see [reporting scalars](../guides/reporting/scalar_reporting.md) / [automatic reporting](../fundamentals/logger.md#automatic-reporting))
* [Plots](#plots)
- Plots are combined to have multiple lines from different experiments (for example multiple RoC curves laid on top
of each other).
* [Debug samples](#debug-samples)
- Debug samples by each iteration
- Examine samples with a viewer (for images and video), and a player (for audio) (see [reporting media](../guides/reporting/media_reporting.md)).
The **ClearML** experiment comparison provides [comparison features](#comparison-features) making it easy to compare experiments.
## Selecting experiments to compare
**To select experiments to compare:**
1. Go to an experiments table, which includes the experiments to be compared.
1. Select the experiments to compare, select the checkboxes individually or select the top checkbox for all experiments. After selecting the second checkbox, a bottom bar appears.
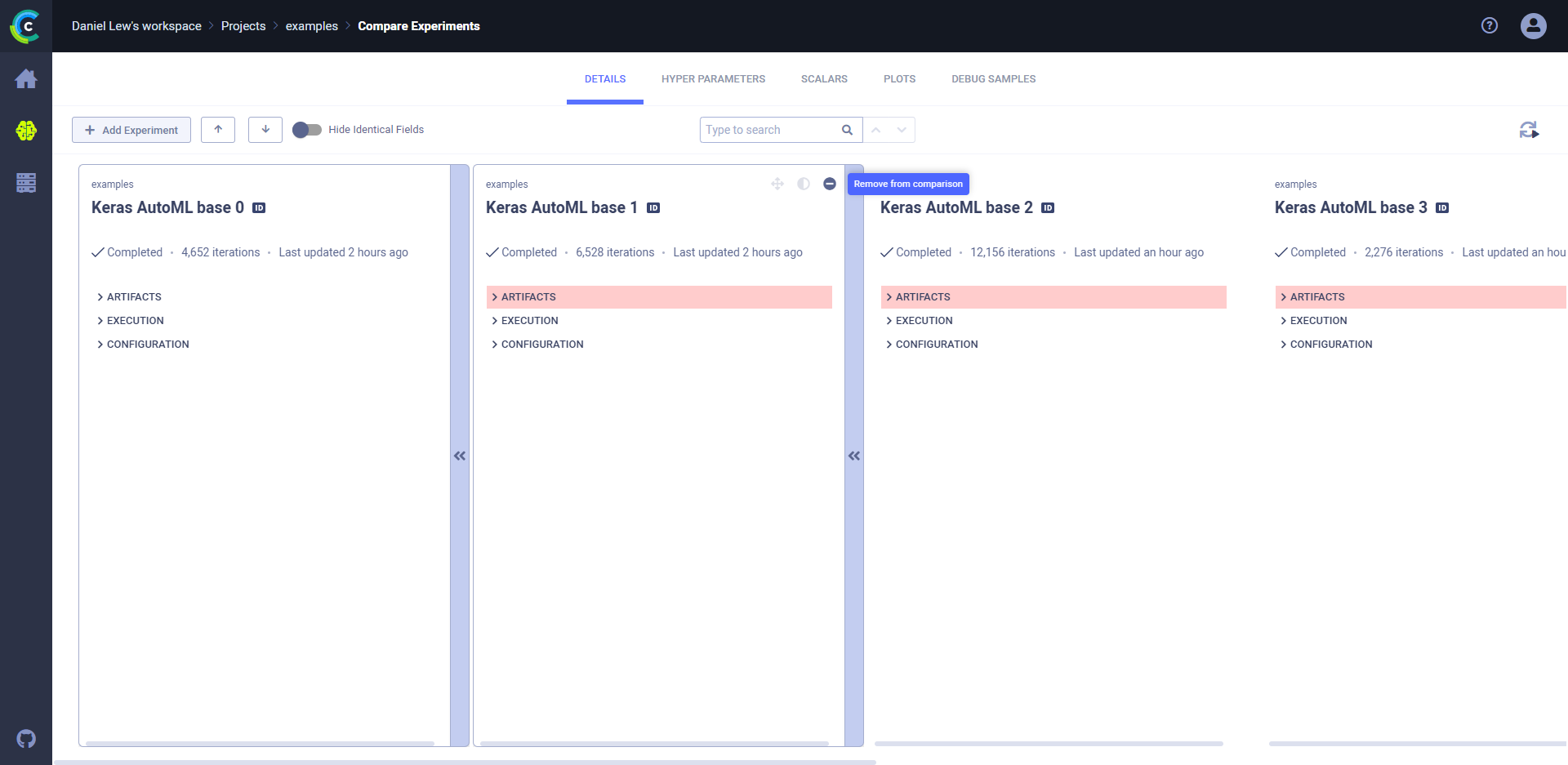
1. In the bottom bar, click **COMPARE**. The comparison page appears, showing a column for each experiment and differences with a highlighted background color. The experiment on the left is the base experiment. Other experiments compare to the base experiment.
## Details
The **DETAILS** tab includes deep comparisons of the following:
### Artifacts
* Input model and model design.
* Output model and model design.
* Other artifacts, if any.
### Execution details
* The Source code - repository, branch, commit ID, script file name, and working directory.
* Uncommitted changes, sorted by file name.
* Installed Python packages and versions, sorted by package name.
### Configuration
* Configuration objects used by the experiment (see [configuration objects](../fundamentals/hyperparameters.md#connecting-objects)),
sorted by sections.
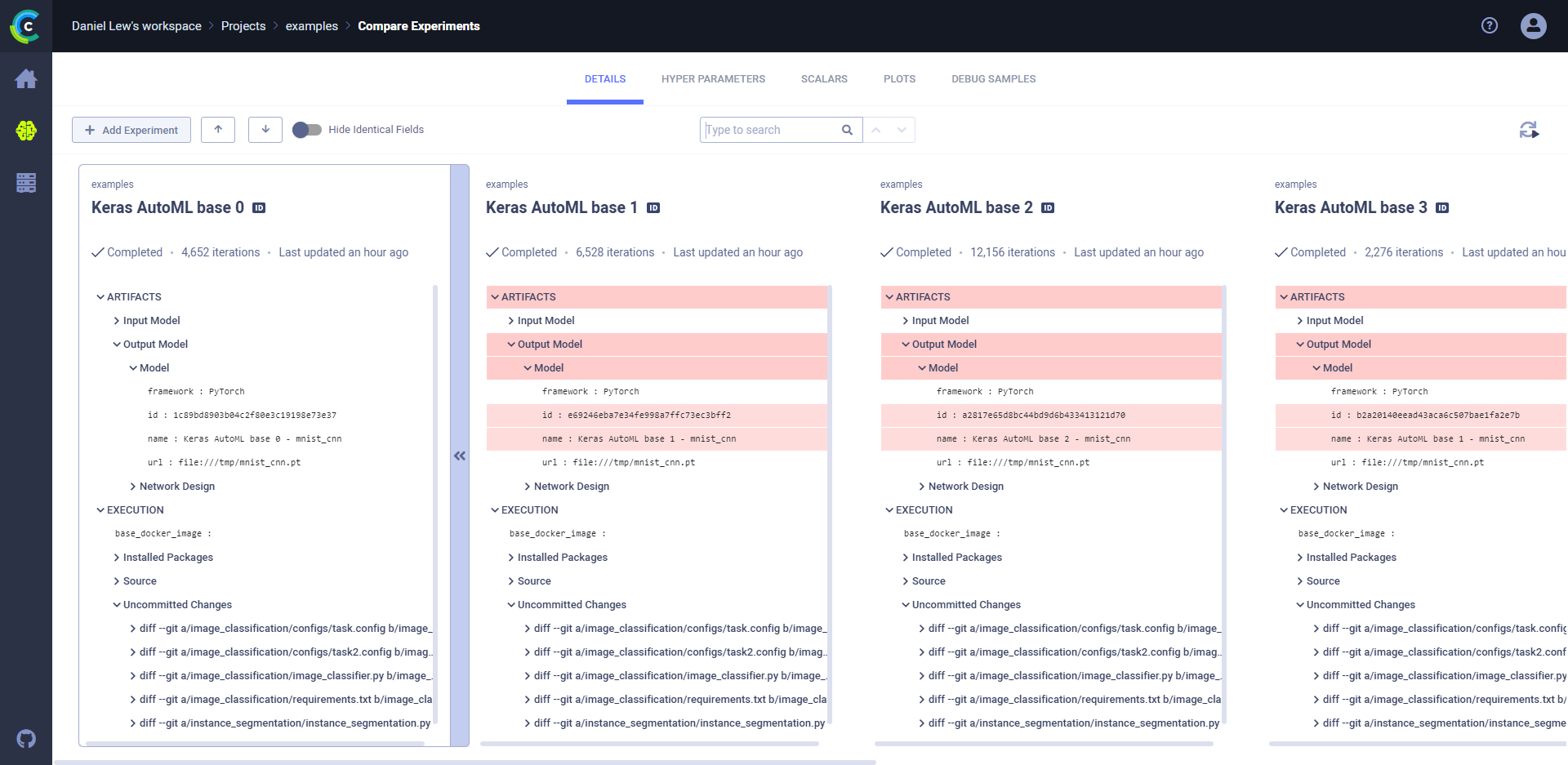
### To locate the source differences:
* Click the **DETAILS** tab **>** Expand highlighted sections, or, in the header, click  (Previous diff) or
(Previous diff) or  (Next diff).
For example, in the image below, expanding **ARTIFACTS** **>** **Output Model** **>** **Model** shows that the model ID
and name are different.
## Hyperparameters
Compare hyperparameters as values, or compare by metric (hyperparameter parallel coordinate comparison).
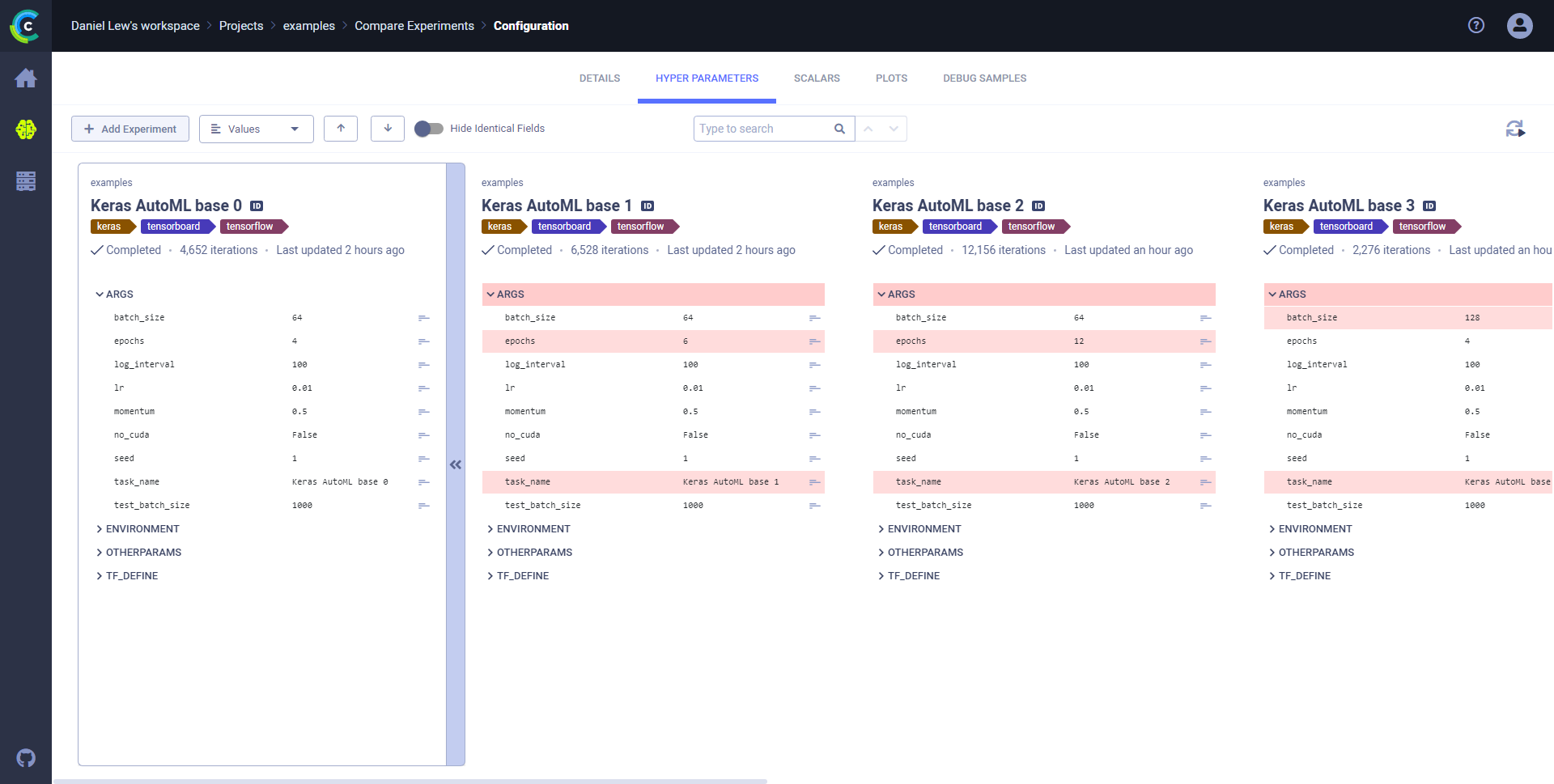
### Values mode
The Values mode is a side-by-side comparison that shows hyperparameter value differences highlighted line-by-line.
**To view a side by side values comparison:**
1. Click the **HYPER PARAMETERS** tab.
1. In the dropdown menu (on the upper left, next to **+ Add Experiments**), choose **Values**.
1. To show only differences, move the **Hide Identical Fields** slider to on.
1. Locate differences by either:
* Clicking
(Next diff).
For example, in the image below, expanding **ARTIFACTS** **>** **Output Model** **>** **Model** shows that the model ID
and name are different.

## Hyperparameters
Compare hyperparameters as values, or compare by metric (hyperparameter parallel coordinate comparison).
### Values mode
The Values mode is a side-by-side comparison that shows hyperparameter value differences highlighted line-by-line.
**To view a side by side values comparison:**
1. Click the **HYPER PARAMETERS** tab.
1. In the dropdown menu (on the upper left, next to **+ Add Experiments**), choose **Values**.
1. To show only differences, move the **Hide Identical Fields** slider to on.
1. Locate differences by either:
* Clicking  (Previous diff) or
(Previous diff) or
 (Next diff).
* Scrolling to see highlighted hyperparameters.
For example, expanding **General** shows that the `batch_size` and `epochs` differ between the experiments.
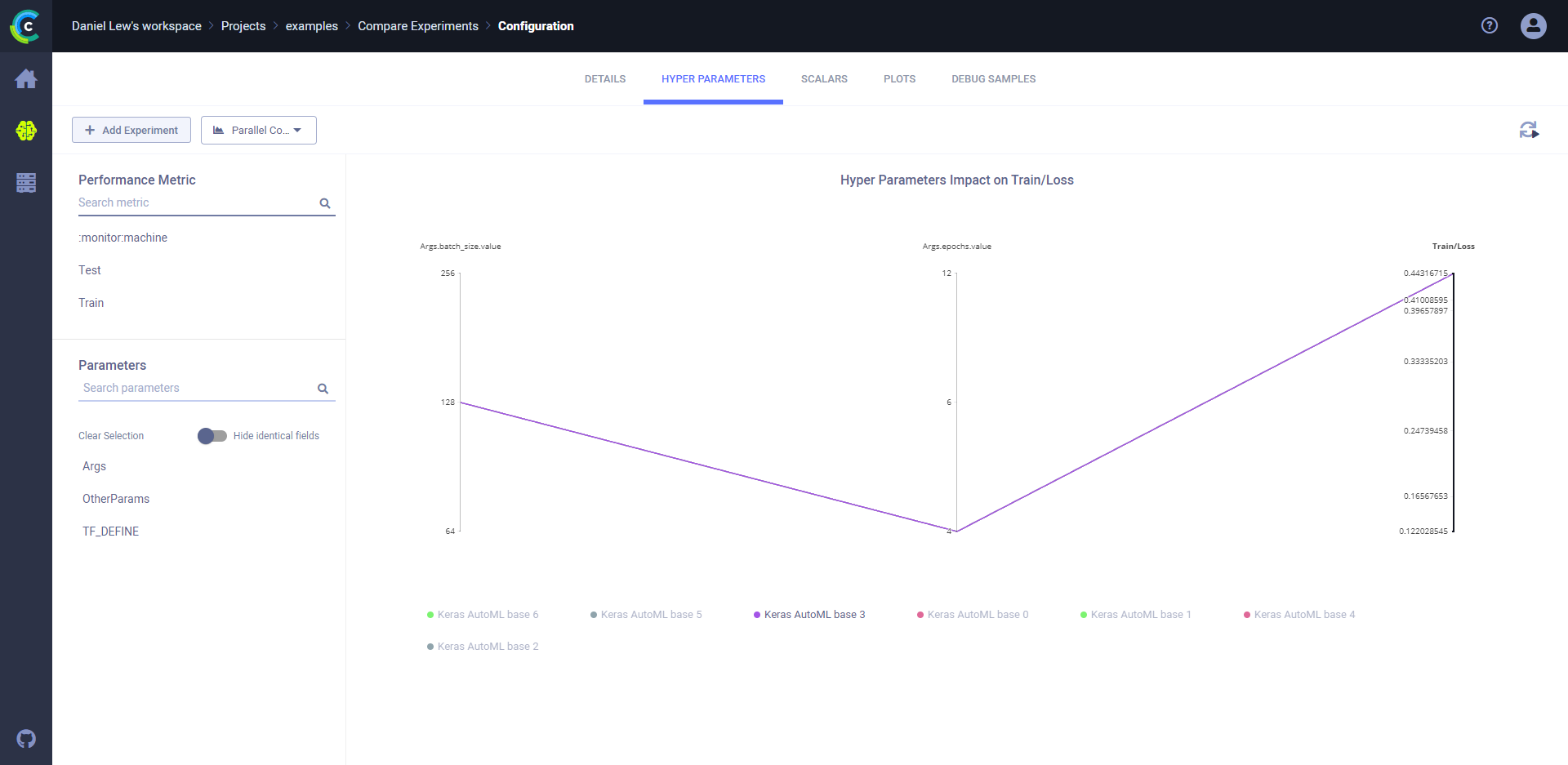
### Parallel Coordinates mode
In the Parallel Coordinates mode, compare a metric to any combination of hyperparameters using a parallel coordinates plot.
**To compare by metric:**
1. Click the **HYPER PARAMETERS** tab.
1. In the dropdown menu (on the upper left, next to **+ Add Experiments**), choose **Parallel Coordinates**.
1. In **Performance Metric**, expand a metric or monitored resource, and then click a variant.
1. Select the metric values to use. Choose one of the following:
* **LAST** - The final value, or the most recent value, for in-progress experiments.
* **MIN** - Minimal value.
* **MAX** - Maximal value.
1. In **Parameters**, select the hyperparameter checkboxes to compare.
1. To view one experiment on the plot, hover over the experiment name in the legend.
For example, plot the metric/variant `epoch_accuracy`/`validation: epoch_accuracy` against the hyperparameters
`batch_size` and `epochs`.

Hover over one of the experiment names in the legend, and the plot shows only that data.
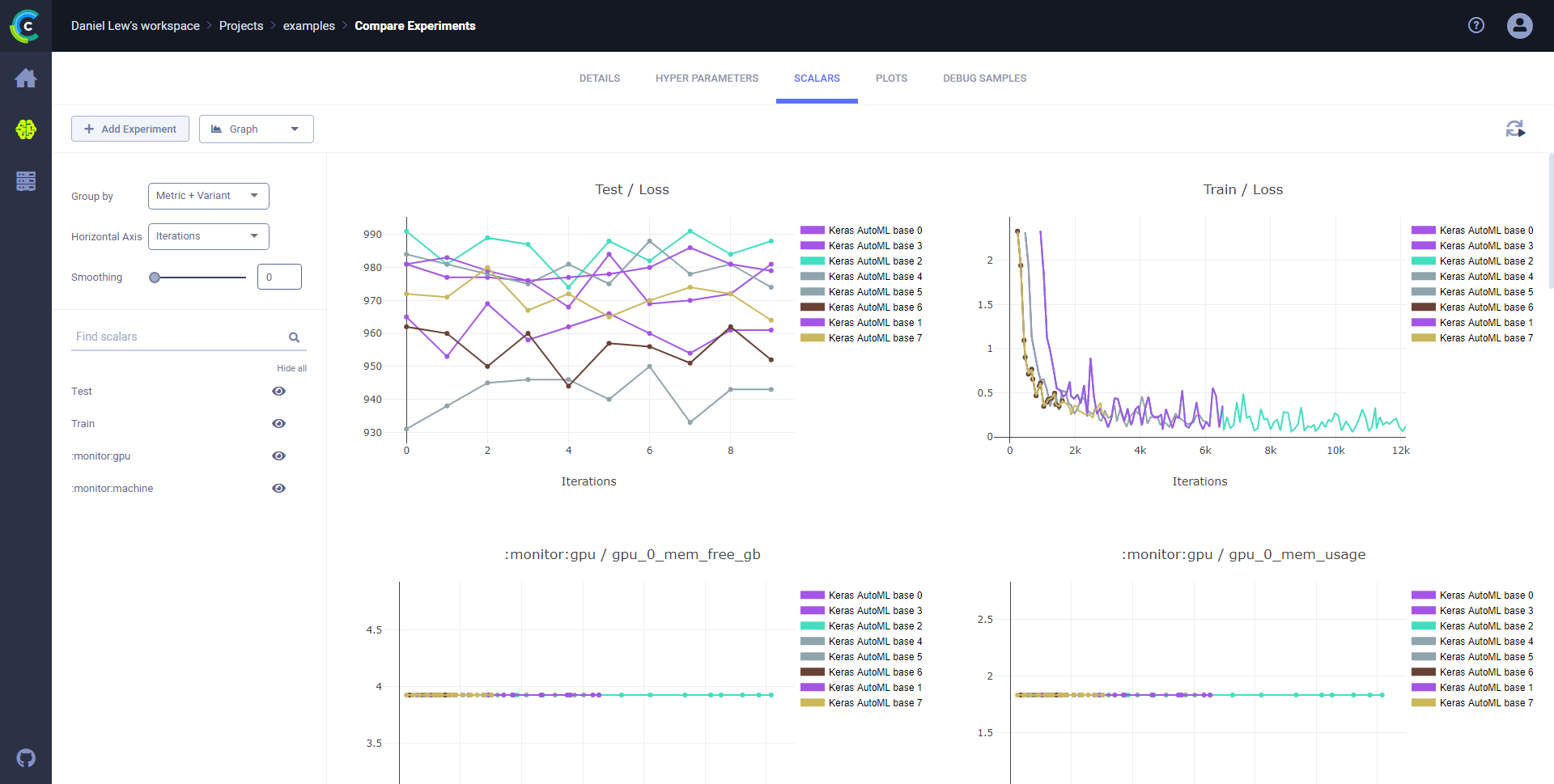
## Scalars
Visualize the comparison of scalars, which includes metrics and monitored resources in the **SCALARS** tab.
### Compare specific values
**To compare specific values:**
1. Click the **SCALARS** tab.
1. In the dropdown menu (upper right of the left sidebar), choose either:
* **Last values** (the final or most recent value)
* **Min Values** (the minimal values)
* **Max Values** (the maximal values)
1. Sort by variant.

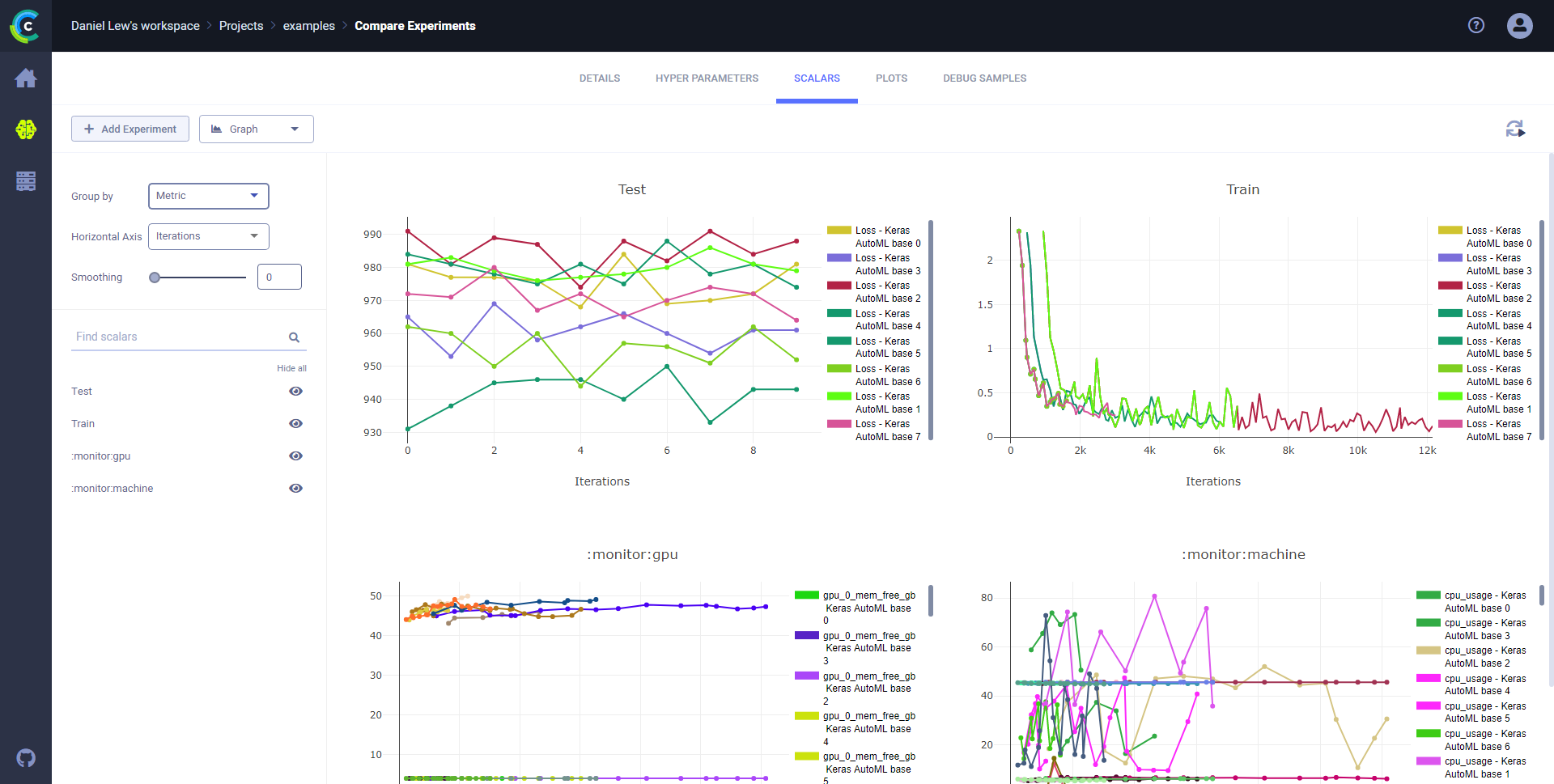
### Compare scalar series
Compare scalar series in plots and analyze differences using **ClearML Web UI** plot tools.
**To compare scalar series:**
1. Click the **SCALARS** tab.
1. In the dropdown menu (upper right of the left sidebar), choose **Graph**.
**To improve scalar series analysis:**
* In **Group by**, select one of these option:
* **Metric** - all variants for a metric on the same plot.
* **Metric+Variant** - every variant appears on its own plot.
* Horizontal axis options:
* Iterations,
* Relative time since the experiment began,
* Wall (clock time).
* Smooth a curve - move the **Smoothing** slider or type in a smoothing number from **0** to **0.999**.
* Use plot controls, which appear when you hover over the top of a plot.
* Hide / show scalar plots - Click **Hide all** and
(Next diff).
* Scrolling to see highlighted hyperparameters.
For example, expanding **General** shows that the `batch_size` and `epochs` differ between the experiments.

### Parallel Coordinates mode
In the Parallel Coordinates mode, compare a metric to any combination of hyperparameters using a parallel coordinates plot.
**To compare by metric:**
1. Click the **HYPER PARAMETERS** tab.
1. In the dropdown menu (on the upper left, next to **+ Add Experiments**), choose **Parallel Coordinates**.
1. In **Performance Metric**, expand a metric or monitored resource, and then click a variant.
1. Select the metric values to use. Choose one of the following:
* **LAST** - The final value, or the most recent value, for in-progress experiments.
* **MIN** - Minimal value.
* **MAX** - Maximal value.
1. In **Parameters**, select the hyperparameter checkboxes to compare.
1. To view one experiment on the plot, hover over the experiment name in the legend.
For example, plot the metric/variant `epoch_accuracy`/`validation: epoch_accuracy` against the hyperparameters
`batch_size` and `epochs`.

Hover over one of the experiment names in the legend, and the plot shows only that data.

## Scalars
Visualize the comparison of scalars, which includes metrics and monitored resources in the **SCALARS** tab.
### Compare specific values
**To compare specific values:**
1. Click the **SCALARS** tab.
1. In the dropdown menu (upper right of the left sidebar), choose either:
* **Last values** (the final or most recent value)
* **Min Values** (the minimal values)
* **Max Values** (the maximal values)
1. Sort by variant.

### Compare scalar series
Compare scalar series in plots and analyze differences using **ClearML Web UI** plot tools.
**To compare scalar series:**
1. Click the **SCALARS** tab.
1. In the dropdown menu (upper right of the left sidebar), choose **Graph**.
**To improve scalar series analysis:**
* In **Group by**, select one of these option:
* **Metric** - all variants for a metric on the same plot.
* **Metric+Variant** - every variant appears on its own plot.
* Horizontal axis options:
* Iterations,
* Relative time since the experiment began,
* Wall (clock time).
* Smooth a curve - move the **Smoothing** slider or type in a smoothing number from **0** to **0.999**.
* Use plot controls, which appear when you hover over the top of a plot.
* Hide / show scalar plots - Click **Hide all** and  .
* Filter scalars by full or partial scalar name.
This image shows scalars grouped by metric.
This image shows scalars grouped by metric and variant.
## Plots
Visualize the comparison of any data that **ClearML** automatically captures or that is explicitly reported in experiments,
in the **PLOTS** tab.
**To compare plots:**
1. Click the **PLOTS** tab.
1. To improve your comparison, use either of the following:
* To locate scalars, click **HIDE ALL**, and then
.
* Filter scalars by full or partial scalar name.
This image shows scalars grouped by metric.

This image shows scalars grouped by metric and variant.

## Plots
Visualize the comparison of any data that **ClearML** automatically captures or that is explicitly reported in experiments,
in the **PLOTS** tab.
**To compare plots:**
1. Click the **PLOTS** tab.
1. To improve your comparison, use either of the following:
* To locate scalars, click **HIDE ALL**, and then  (show) to choose which scalars to see. Scalars can also be filtered by full or partial scalar name, using the search bar.
* Use any of the plot controls that appear when hovering over the top of a plot, including:
* Downloading the image
* Downloading the data as JSON
* Zooming
* Panning
* Switching between logarithmic / linear scale.
## Debug samples
Compare debug samples at any iteration to verify that an experiment is running as expected. The most recent iteration appears
first. Use the viewer / player to inspect images, audio, video samples and do any of the following:
* Move to the same sample in a different iteration (move the iteration slider).
* Show the next or previous iteration's sample.
* Download the file
(show) to choose which scalars to see. Scalars can also be filtered by full or partial scalar name, using the search bar.
* Use any of the plot controls that appear when hovering over the top of a plot, including:
* Downloading the image
* Downloading the data as JSON
* Zooming
* Panning
* Switching between logarithmic / linear scale.
## Debug samples
Compare debug samples at any iteration to verify that an experiment is running as expected. The most recent iteration appears
first. Use the viewer / player to inspect images, audio, video samples and do any of the following:
* Move to the same sample in a different iteration (move the iteration slider).
* Show the next or previous iteration's sample.
* Download the file  .
* Zoom.
* View the sample's iteration number, width, height, and coordinates.
**To compare debug samples:**
1. Click the **DEBUG SAMPLES** tab. The most recent iteration appears at the top.
1. Locate debug samples by doing the following:
* Filter by metric. In the **Metric** list, choose a metric.
* Show other iterations. Click
.
* Zoom.
* View the sample's iteration number, width, height, and coordinates.
**To compare debug samples:**
1. Click the **DEBUG SAMPLES** tab. The most recent iteration appears at the top.
1. Locate debug samples by doing the following:
* Filter by metric. In the **Metric** list, choose a metric.
* Show other iterations. Click  (Older images),
(Older images),
 (New images), or
(New images), or  (Newest images).

1. To open a debug sample (image, audio, or video) in the viewer or player, click the thumbnail.

1. To move to the same sample in another iteration, click
(Newest images).

1. To open a debug sample (image, audio, or video) in the viewer or player, click the thumbnail.

1. To move to the same sample in another iteration, click  (previous),
(previous),  (next), or move the slider.
**To view a debug sample in the viewer / player:**
1. Click the debug sample click the thumbnail.
1. Do any of the following:
* Move to the same sample in another iteration - Move the slider, or click **<** (previous) or **>** (next).
* Download the file - Click
(next), or move the slider.
**To view a debug sample in the viewer / player:**
1. Click the debug sample click the thumbnail.
1. Do any of the following:
* Move to the same sample in another iteration - Move the slider, or click **<** (previous) or **>** (next).
* Download the file - Click  .
* Zoom
* For images, locate a position on the sample - Hover over the sample and the X, Y coordinates appear in the legend below the sample.
## Comparison features
To assist in experiment analysis, the comparison page supports:
* [Adding experiments to the comparison](#adding-experiments-to-the-comparison) using a partial name search.
* [Finding the next or previous difference](#finding-the-next-or-previous-difference).
* [Hiding identical fields](#hiding-identical-fields)
* [Searching all text](#searching-all-text)
* [Choosing a different base experiment](#choosing-a-different-base-experiment)
* [Dynamic ordering](#dynamic-ordering-of-the-compared-experiments) of the compared experiments
* [Sharing experiments](#sharing-experiments)
* Auto refresh
### Adding experiments to the comparison
Add an experiment to the comparison - Click **Add Experiment** and start typing an experiment name. An experiment search
and select dialog appears showing matching experiments to choose from. To add an experiment, click **+**. To Remove
an experiment, click
.
* Zoom
* For images, locate a position on the sample - Hover over the sample and the X, Y coordinates appear in the legend below the sample.
## Comparison features
To assist in experiment analysis, the comparison page supports:
* [Adding experiments to the comparison](#adding-experiments-to-the-comparison) using a partial name search.
* [Finding the next or previous difference](#finding-the-next-or-previous-difference).
* [Hiding identical fields](#hiding-identical-fields)
* [Searching all text](#searching-all-text)
* [Choosing a different base experiment](#choosing-a-different-base-experiment)
* [Dynamic ordering](#dynamic-ordering-of-the-compared-experiments) of the compared experiments
* [Sharing experiments](#sharing-experiments)
* Auto refresh
### Adding experiments to the comparison
Add an experiment to the comparison - Click **Add Experiment** and start typing an experiment name. An experiment search
and select dialog appears showing matching experiments to choose from. To add an experiment, click **+**. To Remove
an experiment, click  .

### Finding the next or previous difference
* Find the previous difference
.

### Finding the next or previous difference
* Find the previous difference  , or
the next difference
, or
the next difference  .
### Hiding identical fields
Move the **Hide Identical Fields** slider to "on" mode to see only fields that are different.
### Searching all text
Search all text in the comparison.
### Choosing a different base experiment
Show differences in other experiments in reference to a new base experiment. To set a new base experiment, do one of the following:
* Click on
.
### Hiding identical fields
Move the **Hide Identical Fields** slider to "on" mode to see only fields that are different.
### Searching all text
Search all text in the comparison.
### Choosing a different base experiment
Show differences in other experiments in reference to a new base experiment. To set a new base experiment, do one of the following:
* Click on  on the top right of the experiment that will be the new base.
* Click on
on the top right of the experiment that will be the new base.
* Click on  the new base experiment and drag it all the way to the left

### Dynamic ordering of the compared experiments
To reorder the experiments being compared, press
the new base experiment and drag it all the way to the left

### Dynamic ordering of the compared experiments
To reorder the experiments being compared, press  on the top right of the experiment that
needs to be moved, and drag the experiment to its new position.

### Removing an experiment from the comparison
Remove an experiment from the comparison, by pressing
on the top right of the experiment that
needs to be moved, and drag the experiment to its new position.

### Removing an experiment from the comparison
Remove an experiment from the comparison, by pressing  on the top right of the experiment that needs to be removed.
### Sharing experiments
To share a comparison table, copy the full URL from the address bar and send it to a teammate to collaborate. They will
get the exact same page (including selected tabs etc.).
on the top right of the experiment that needs to be removed.

### Sharing experiments
To share a comparison table, copy the full URL from the address bar and send it to a teammate to collaborate. They will
get the exact same page (including selected tabs etc.). .
* Zoom.
* View the sample's iteration number, width, height, and coordinates.
**To compare debug samples:**
1. Click the **DEBUG SAMPLES** tab. The most recent iteration appears at the top.
1. Locate debug samples by doing the following:
* Filter by metric. In the **Metric** list, choose a metric.
* Show other iterations. Click
.
* Zoom.
* View the sample's iteration number, width, height, and coordinates.
**To compare debug samples:**
1. Click the **DEBUG SAMPLES** tab. The most recent iteration appears at the top.
1. Locate debug samples by doing the following:
* Filter by metric. In the **Metric** list, choose a metric.
* Show other iterations. Click  (Older images),
(Older images),
 (New images), or
(New images), or  (Newest images).

1. To open a debug sample (image, audio, or video) in the viewer or player, click the thumbnail.

1. To move to the same sample in another iteration, click
(Newest images).

1. To open a debug sample (image, audio, or video) in the viewer or player, click the thumbnail.

1. To move to the same sample in another iteration, click  .
* Zoom
* For images, locate a position on the sample - Hover over the sample and the X, Y coordinates appear in the legend below the sample.
## Comparison features
To assist in experiment analysis, the comparison page supports:
* [Adding experiments to the comparison](#adding-experiments-to-the-comparison) using a partial name search.
* [Finding the next or previous difference](#finding-the-next-or-previous-difference).
* [Hiding identical fields](#hiding-identical-fields)
* [Searching all text](#searching-all-text)
* [Choosing a different base experiment](#choosing-a-different-base-experiment)
* [Dynamic ordering](#dynamic-ordering-of-the-compared-experiments) of the compared experiments
* [Sharing experiments](#sharing-experiments)
* Auto refresh
### Adding experiments to the comparison
Add an experiment to the comparison - Click **Add Experiment** and start typing an experiment name. An experiment search
and select dialog appears showing matching experiments to choose from. To add an experiment, click **+**. To Remove
an experiment, click
.
* Zoom
* For images, locate a position on the sample - Hover over the sample and the X, Y coordinates appear in the legend below the sample.
## Comparison features
To assist in experiment analysis, the comparison page supports:
* [Adding experiments to the comparison](#adding-experiments-to-the-comparison) using a partial name search.
* [Finding the next or previous difference](#finding-the-next-or-previous-difference).
* [Hiding identical fields](#hiding-identical-fields)
* [Searching all text](#searching-all-text)
* [Choosing a different base experiment](#choosing-a-different-base-experiment)
* [Dynamic ordering](#dynamic-ordering-of-the-compared-experiments) of the compared experiments
* [Sharing experiments](#sharing-experiments)
* Auto refresh
### Adding experiments to the comparison
Add an experiment to the comparison - Click **Add Experiment** and start typing an experiment name. An experiment search
and select dialog appears showing matching experiments to choose from. To add an experiment, click **+**. To Remove
an experiment, click  the new base experiment and drag it all the way to the left

### Dynamic ordering of the compared experiments
To reorder the experiments being compared, press
the new base experiment and drag it all the way to the left

### Dynamic ordering of the compared experiments
To reorder the experiments being compared, press  on the top right of the experiment that
needs to be moved, and drag the experiment to its new position.

### Removing an experiment from the comparison
Remove an experiment from the comparison, by pressing
on the top right of the experiment that
needs to be moved, and drag the experiment to its new position.

### Removing an experiment from the comparison
Remove an experiment from the comparison, by pressing